
Associate Professor & Head, Department of Computational Sciences, School of Basic Sciences
- Ph.D. Crystallography and Biophysics, Centre of Advanced Study in Crystallography and Biophysics, University of Madras, Chennai, India (2006)
- M.Sc. Physics, N.G.M. College, Bharathiar University, Coimbatore, India (1999)
- B.Sc. Physics, N.G.M. College, Bharathiar University, Coimbatore, India (1997)
Teaching and Research Experience
| Institute | Position | Experience | Period |
|---|---|---|---|
| Department of Computational Sciences, Central University of Punjab | Associate Professor | Teaching and Research | Jan 2022 - Present |
| Department of Bioinformatics, School of Chemical & Biotechnology, SASTRA Deemed University | Associate Professor | Teaching and Research | Sep 2019 - Jan 2022 |
| Department of Bioinformatics, School of Chemical & Biotechnology, SASTRA Deemed University | Senior Assistant Professor | Teaching and Research | June 2012 -Aug 2019 |
| Department of Life Sciences, Shiv Nadar University | Assistant Professor | Teaching | Jan 2012 - May 2012 |
| Molecular Cell Biology Unit, Dept. of Biology, Lund University, Sweden | Post-Doctoral Research Scientist | Research | Jan 2011 - Dec 2011 |
| Center for Molecular Protein Science, Lund University, Sweden. | Post-Doctoral Research Scientist | Research | Sep 2006 - Dec 2010 |
Administrative Experience
Mar 2022 - Present : Head of the Department of Computational Sciences, Central University of Punjab
Any Other Experience
Associate Editor - PLOS One, BMC Research Notes, Current Bioinformatics, Frontiers in Molecular Biosciences, Current Indian Science
Structural Biology, Macromolecular Crystallography, Protein Biophysics, Structural Bioinformatics, Structural & Functional genomics, Sequence data analysis.
- Structure-function relationship of proteins of biological and medicinal interest using structural & biophysical methods.
- Studying Protein-ligand recognition using X-ray and neutron crystallography and computational biology.
- Protein folding / misfolding and oligomerisation using Biophysical, Structural biology and Bioinformatics approach.
- Mutation-induced changes in structural stability, folding and dynamics of medically important proteins.
- Structure-based drug design for human disease and infections using structural and computational biology methods.
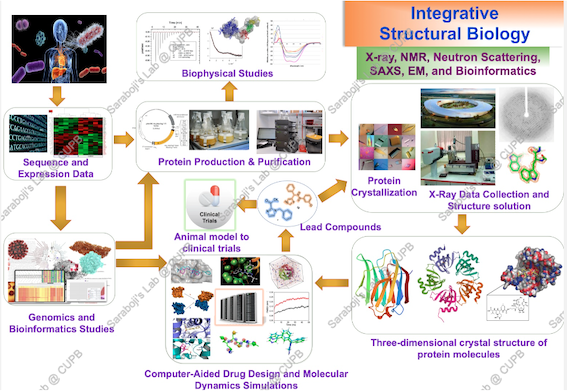
- Structural and biophysical studies on human Cu/Zn SOD1 mutants associated with the motor neuron disease, ALS: a promising model system for understanding the origin of protein-misfolding disease and structure-based design of novel inhibitors against SOD1 misfolding and aggregation. Approved by ICMR, Govt. of India [2022].
- Structural, biochemical and computational investigation on the therapeutic targets of SARS-CoV-2 and structure-based drug discovery. Funded by SERB-COVID-19 special call, Govt. of India [2020-2021].
- Structural studies on the antioxidant enzymes of Wuchereria bancrofti towards the development of novel structure based drug lead discovery for Lymphatic filariasis. Funded by SERB, Govt. of India [2018-2022].
- Structural studies on disease triggering mutations of superoxide dismutase (SOD1), located far from the active site and dimer interface. Funded by SERB, Govt. of India [2013-2016] .
- Purification, Crystallization and Structural Studies of Hemoglobin From species living in extreme conditions. Funded by SASTRA-TRR research grant [2013-2014].
- Indian Crystallographic Association (ICA)
- Indian Biophysical Society (IBS)
- The Indian Science Congress Association (ISCA)
- The Society of Biological Chemists INDIA (SBC[I])
- Indian Society of Systems for Science and Engineering (ISSE)
- International Society for Computational Biology (ISCB)
- Post-Doctoral Fellowship, Dept. of Biology, Lund University, Sweden (Jab 2011- Jan 2012).
- Post-Doctoral Fellowship, Dept. Biochemistry and Structural Biology, Lund University, Sweden. (Apr 2009 – Dec 2010).
- Post-Doctoral Fellowship, Dept. Biochemistry and Biophysics, Stockholm University, Sweden. (Sep 2006 - Mar 2009).
- Senior Research Fellowship, Council of Scientific and Industrial Research, Govt. of India. (2004-2006).
- Prof. M. Michael Gromiha, Indian Institute of Technology-Madras.
- Prof. J. Jeyakanthan, Alagappa University, Karaikudi.
- Dr. A. S. Ethayathulla, All India Institute of Medical Sciences, New Delhi.
- Dr. R. Thenmalarchelvi, Indian Institute of Technology-Hyderabad.
- Dr. D. Bharanidharan, Aravind Medical Research Foundation, Madurai.
- Sureshan, M., Prabhu, D., and Rajamanikandan S., Saraboji, K. (2023) Discovery of potent inhibitors targeting Glutathione S-transferase of Wuchereria bancrofti: a step toward the development of effective anti-filariasis drugs. Molecular Diversity (Online ahead of print).
- Sureshan, M., Prabhu, D., and Saraboji, K. (2022) Computational identification and experimental validation of anti-filarial lead molecules targeting metal binding/substrate channel residues of Cu/Zn SOD1 from Wuchereria bancrofti. J Biomol Struct Dyn. (Online ahead of print.)
- Sureshan, M., Rajamanikandan S, Srimari, S., Prabhu, D., Jeyakanthan J. and Saraboji, K. (2022) Designing specific inhibitors against Dihydrofolate reductase of W. bancrofti towards drug discovery for lymphatic filariasis. Structural Chemistry, 33, 935–947.
- Srimari S., Thamarai Selvi, S., Prabhu, D. and Saraboji K. (2021) Advances in the computational methods to understand protein folding and stability. Int. J. Data Min. Bioinform. 26, 202-223.
- Sureshan, M., Prabhu, D., Aruldoss, I., and Saraboji, K. (2021) Potential inhibitors for peroxiredoxin 6 of W. bancrofti: A combined study of modelling, structure-based drug design and MD simulation. J. Mol. Graph. Model. 112, 108115.
- Prabhu D, Rajamanikandan S, Sureshan M, Jeyakanthan J, and Saraboji K. (2021) Modelling studies reveal the importance of the C-terminal inter motif loop of NSP1 as a promising target site for drug discovery and screening of potential phytochemicals to combat SARS-CoV-2. J. Mol. Graph. Model. 106, 107920.
- Dharma Rao, T, Aruldoss, I., Srimari, S., and Saraboji, K. (2021) Trends and strategies to combat viral infections: a review on FDA approved antiviral drugs. Int. J. Biol. Macromol. 172, 524-541.
- Dharma Rao, T, Sureshan, M., Srimari, S., and Saraboji, K. (2020) Molecular dynamics of far positioned surface mutations of Cu/Zn SOD1 promotes altered structural stability and metal-binding site: Structural clues to the pathogenesis of Amyotrophic Lateral Sclerosis. J. Mol. Graph. Model. 100, 107678.
- Dharma Rao, T and Saraboji, K. (2020) Changes in hydrophobicity mainly promotes the aggregation tendency of ALS associated SOD1 mutants. Int J Biol Macromol. 145, 904-913.
- Sarwareddy, K., Sureshan, M., Saraboji. K., Krishna Priya. M. (2020) In-silico and in-vitro analysis of endocan interaction with statins. Int J Biol Macromol.146, 1087-1099.
- Thiagarajan, R., Varsha, MKNS., Srinivasan, V., Ravichandran, R., Saraboji, K. (2019) Vitamin K1 prevents diabetic cataract by inhibiting lens aldose reductase 2 (ALR2) activity. Sci Rep. 9, 14684.
- Dharma Rao, T and Saraboji, K. (2019) Far positioned ALS associated mutants of Cu/Zn SOD forms partially metallated, destabilized misfolding intermediates. Biochem. Biophys. Res. Commun., 516, 494-499.
- Dharma Rao, T and Saraboji, K. (2018) Molecular dynamics of a far positioned SOD1 mutant V14M reveals pathogenic misfolding behavior. J Biomol Struct Dyn. 36, 4085-4098.
- Sureshkumar, K., Maheshwaran, V., Dharma Rao, T., Themmila, K., Ponnuswamy, M.N., Saraboji, K., Dhandayutham, S. (2017) Synthesis, characterization, crystal structure, in-vitro anti-inflammatory and molecular docking studies of 5-mercapto-1-substituted tetrazole incorporated quinoline derivative. J. Mol. Struct. 1146, 314-323.
- Manzoni, F., Saraboji, K., Sprenger, J., Kumar, R., Noresson. A.L., Nilsson, U.J., Leffler, H., Fisher, S.Z., Schrader, T.E., Ostermann, A., Coates, L., Blakeley, M.P., Oksanen, E. and Logan, D.T. (2016) Perdeuteration, crystallization, data collection and comparison of five neutron diffraction data sets of complexes of human galectin-3C. Acta Cryst. Sec D., 72, 1194-1202.
- Tompa, D.R., Gromiha M, M., and Saraboji, K. (2016) Contribution of main chain and side chain atoms and their locations to the stability of thermophilic proteins. J. Mol. Graph. Model. 64, 85-93.
- Shaistha, T and Saraboji, K. (2015) Analysis of beta-structure interfaces of various protein folds. J. Comput. Theor. Nanosci., 12, 2237-2245.
- Abubakkar, M.M, Saraboji, K and Ponnuswamy, M.N. (2014) The Crystal Structure of Oxy Hemoglobin from High Oxygen Affinity Bird Emu. Gen. Physiol. Biophys., 33, 373-382.
- Danielsson, J., Awad, W., Saraboji, K., Kurnik, M., Lang, L., Leinartaité, L., Marklund, S.L., Logan, D.T. and Oliveberg, M. (2013) Global structural motions from the strain of a single hydrogen bond. Proc. Natl. Acad. Sci. USA. 110, 3829-3834.
- Gromiha, M.M., Pathak, M.C., Saraboji, K., Ortlund, E.A., and Gaucher, E.A. (2013). Hydrophobic environment is a key factor for the stability of thermophilic proteins. Proteins: Struct. Funct. Bioinf. 81, 715-721.
- Mohamed Abubakkar, M., Saraboji, K., and Ponnuswamy, M.N (2013). Purification, crystallization and preliminary crystallograpic studies of haemoglobin from mongoose (Helogale parvula) in two different crystal forms induced by pH variation. Acta Cryst., F69, 126–129.
- Saraboji, K., Håkansson, M., Genheden, S., Diehl, C., Qvist, J., Weininger, U., Nilsson,U., Leffler, H., Ryde, U., Akke, M. and Logan, D.T. (2012). The carbohydrate-binding site in galectin-3 is pre-organized to recognize a sugar-like framework of oxygens: ultra-high-resolution structures and water dynamics. Biochemistry, 51, 296-306.
- Haglund, E., Danielsson, J., Saraboji, K., Lindberg, M.O, Logan, D.T., Oliveberg, M. (2012) Trimming down a protein structure to its bare foldons: spatial organization of the cooperative unit. J. Biol. Chem. 287, 2731-2738.
- Leinartaité, L., Saraboji, K., Logan, D.T. and Oliveberg, M. (2010). Folding catalysis by transient coordination of Zn2+ to the Cu ligands of the ALS-associated enzyme Cu/Zn Superoxide dismutase 1. J. Am. Chem. Soc., 132, 13495-13504.
- Nordlund, A*., Leinartaité, L*., Saraboji, K*., Aisenbrey,C., Gröbner, G., Danielsson, J., Logan, D.T. and Oliveberg, M. (2009). Functional features cause misfolding of the ALS-provoking enzyme SOD1. Proc. Natl. Acad. Sci. USA. 106, 9667. (*Equal contribution).
- Mizutani, H., Saraboji, K., Malathy Sony, S.M., Ponnuswamy, M.N., Kumarevel, T., Krishna Swamy, B.S., Simanshu, D.K., Murthy, M.R.N. and Kunishimaa, N. (2008). Systematic study on crystal-contact engineering of diphthine synthase: influence of mutations at crystal-packing regions on X-ray diffraction quality. Acta Cryst., D64, 1020.
- Huang, L-T., Saraboji, K., Ho, S-Y., Hwang, S-F., Ponnuswamy, M.N. and Gromiha, M.M. (2007). Prediction of protein mutant stability using classification and regression tool. Biophys. Chem., 125, 462.
- Balasundaresan, D., Saraboji, K. and Ponnuswamy, M.N. (2006). Crystal structure of haemoglobin from donkey (Equus asinus) at 3Å resolution. Biochimie, 88, 719-723.
- Saraboji, K., Gromiha, M.M. and Ponnuswamy, M.N. (2006). Average assignment method for predicting the stability of protein mutants. Biopolymers, 82, 80-92.
- Malathy Sony, S.M., Saraboji, K., Sukumar, N. and Ponnuswamy, M.N. (2006). Role of amino acid properties to determine backbone t (N-Ca-C') stretching angle in peptides and proteins. Biophys. Chem., 120, 24-31.
- Ponnuswamy, M.N., Gromiha, M.M., Malathy Sony, S.M. and Saraboji, K. (2006). Conformational aspects and interaction studies of heterocyclic drugs. “QSAR and Molecular Modeling of Heterocyclic Drugs”, 3, 81-147. (Ed. S.P. Gupta), Springer Publishers, Germany. (Book Chapter)
- Saraboji, K., Gromiha, M.M. and Ponnuswamy, M.N. (2005). Relative importance of secondary structure and solvent accessibility to the stability of protein mutants: A case study with amino acid properties and energetics on T4 and human lysozymes. Comp. Biol. Chem., 29, 25-35.
- Saraboji, K., Gromiha, M.M. and Ponnuswamy, M.N. (2005). Importance of main-chain hydrophobic free energy to the stability of thermophilic proteins. Int. J. Biol. Macromol., 35, 211.
- Nallini, A., Saraboji, K., Ponnuswamy, M.N., Venkatraj, M. and Jeyaraman, R. (2005). Crystal structure of N1,N5-dibenzoyl tetra hydro -4-methyl-1,5-benzodiazepin-2-one (DBTBO) and tetrahydro-4-methyl-one,5-benzodiazepin-2-one (TBO). Cryst. Res. Technol., 40, 622-626.
- Balasundaresan, D., Saraboji, K and Ponnuswamy, M.N. (2004). Purification and crystallization of haemoglobin from donkey. Biochem. Biophys. Res. Commun., 313, 466-467.
- Gromiha, M.M., Saraboji, K., Ahmad, S., Ponnuswamy, M.N. and Suwa, M. (2004). Role of non-covalent interactions for determining the folding rate of two-state proteins. Biophys. Chem.,107, 263.
- Nallini, A., Saraboji, K., Ponnuswamy, M.N. and Katti, S.B. (2004). Crystallization and structural studies of N-benzyl oxycarbonyl-L-Tyrosine-L-Glysine- Ethyl Ester (Z-Tyr-Gly-OEt). Cryst. Res. Technol., 39, 279-286.
- Malathy Sony, S.M., Saraboji, K., Ponnuswamy, M.N., Manaonmani, J., Kandasamy, M. and Fun, H.K. (2004). Structure and conformation of a nickel complex: {2-Hydroxo-3-piperidine-1-ylmethyl-N, N'(bis-5-promobenzylpropylenediimine) nickel (II) perchlorate}. Cryst. Res. Technol., 39,185.
- Nallini, A., Saraboji, K., Ponnuswamy, M.N. and Katti, S.B. (2004). Structure and conformation of N-t-Butoxycarbonyl-L-valine-L-phenylaline methyl ester (Boc-Val-Phe-OMe). Cryst. Res. Technol., 39, 179-184.
- Nallini, A., Saraboji, K. and Ponnuswamy, M.N. (2004). A study of aromatic hydrogen bonds for peptides with aromatic amino acid side-chains. Ind. J. Biochem. Biophys., 41, 184-187.
- Nallini, A., Saraboji, K., Ponnuswamy, M.N., Muthukumar, M. and Jeyaraman, R. (2003). Crystal structures of pair of benzothiazepines. Mol.Cryst. Liq.Cryst., 393, 83-93.
- Nallini,A., Saraboji, K., Ponnuswamy, M.N., Venkatraj, M. and Jeyaraman, R. (2003). Crystal structure and conformation of two similar piperidones. Mol. Cryst. Liq. Cryst., 403, 49-56.
- Nallini, A., Saraboji, K., Ponnuswamy, M.N., Venkatraj, M. and Jeyaraman, R. (2003). Crystal structure and conformation of a pair of piperidine derivatives. Mol.Cryst.Liq.Cryst., 403, 57-65.
Books Authored:
Saraboji. K., Dharma Rao. T., Prabhu, D. (2022) Promising Antiviral Potentials of Phytochemicals, LAP LAMBERT Academic Publishing.
Book Chapters:
Ponnuswamy, M.N., Gromiha, M.M., Malathy Sony, S.M. and Saraboji, K. (2006). Conformational aspects and interaction studies of heterocyclic drugs. “QSAR and Molecular Modeling of Heterocyclic Drugs”, 3, 81-147. (Ed. S.P. Gupta), Springer Publishers, Germany.
Ph.D. Supervision:
Dr. Dharma Rao T (Completed in 2019; Currently @ Institute of Life Sciences, Bhubaneswar)
Mr. Sureshan M (On Going)
Ms. Priyanka J (On Going)
Ph.D. Co-Supervision:
Dr. Suganya S (Completed in 2019; Currently @ Northwestern University, Chicago)
M.Tech. / M.Sc. projects - 14 candidates
International Level
- Convener, International Conference on Synergy of Sciences (ICSS-2020), Feb 27-29, 2020, SASTRA University. (Supported by SERB, DBT, CSIR)
- Convener, 2nd International Conference on Structural & Functional Genomics (ICSAFG-2016), August 19-20, 2016, SASTRA University. (Supported by SERB, DBT, ICMR)
- Convener, International Conference on Crystal Growth and Biomolecular Crystallography (ICCGBC-2014), November 28-29, 2014, SASTRA University. (Supported by SERB, DBT, CSIR)
National Level
- Symposium on Data Analytics in Bioinformatics, 3 Feb 2018, SASTRA University
- Symposium on Modern Biology, 17th Oct 2015, SASTRA University
- Schrodinger Workshop on Computational Drug Design, March 27-28, 2015, SASTRA University
- Application of X-ray Crystallography for Three-dimensional Structure Determination, Nov 27, 2014, SASTRA University
Macromolecular Structures Deposited in RCSB-Protein Data Bank (PDB) ( https://www.rcsb.org ) are Available here
https://sites.google.com/cup.edu.in/saraboji-pbd/home
